Vizualise how missing data thresholds affect sample clustering
assess_missing_data_tsne.RdThis function can be run in two ways: 1) Without 'thresholds' specified. This will run t-SNE for the input vcf without filtering, and visualize the clustering of samples in two-dimensional space, coloring each sample according to a priori population assignment given in the popmap. 2) With 'thresholds' specified. This will filter your input vcf file to the specified missing data thresholds, and run a t-SNE clustering analysis for each filtering iteration. For each iteration, a 2D plot will be output showing clustering according to the specified popmap. This option is ideal for assessing the effects of missing data on clustering patterns.
assess_missing_data_tsne(
vcfR,
popmap = NULL,
thresholds = NULL,
perplexity = NULL,
iterations = NULL,
initial_dims = NULL,
clustering = TRUE
)Arguments
- vcfR
a vcfR object
- popmap
set of population assignments that will be used to color code the plots
- thresholds
a vector specifying the missing data filtering thresholds to explore
- perplexity
numerical value specifying the perplexity paramter during t-SNE (default: 5)
- iterations
a numerical value specifying the number of iterations for t-SNE (default: 1000)
- initial_dims
a numerical value specifying the number of initial_dimensions for t-SNE (default: 5)
- clustering
use partitioning around medoids (PAM) to do unsupervised clustering on the output? (default = TRUE, max clusters = # of levels in popmap + 2)
Value
a series of plots showing the clustering of all samples in two-dimensional space
Examples
assess_missing_data_tsne(vcfR = SNPfiltR::vcfR.example,
popmap = SNPfiltR::popmap,
thresholds = .8)
#> cutoff is specified, filtered vcfR object will be returned
#> 65.8% of SNPs fell below a completeness cutoff of 0.8 and were removed from the VCF
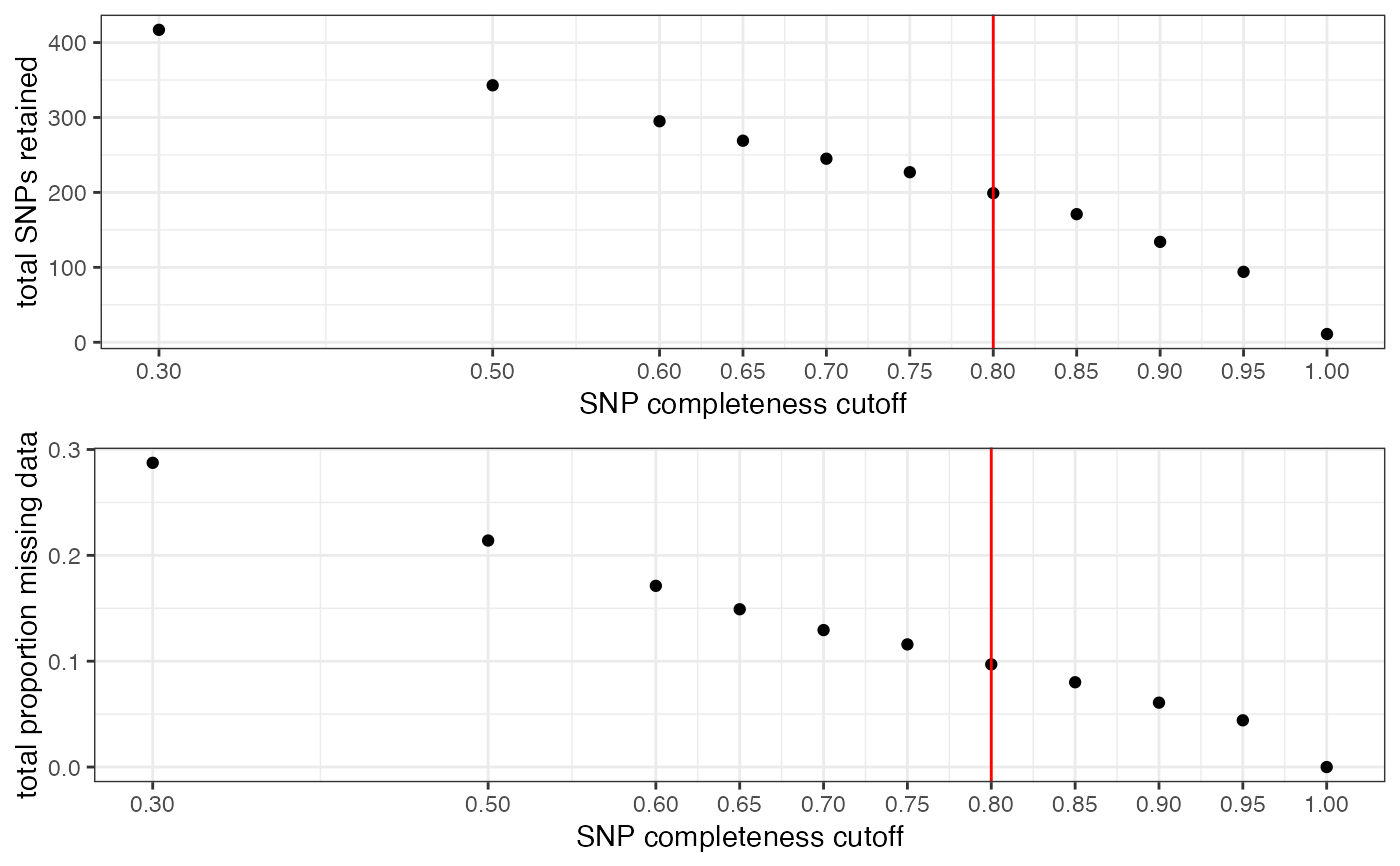
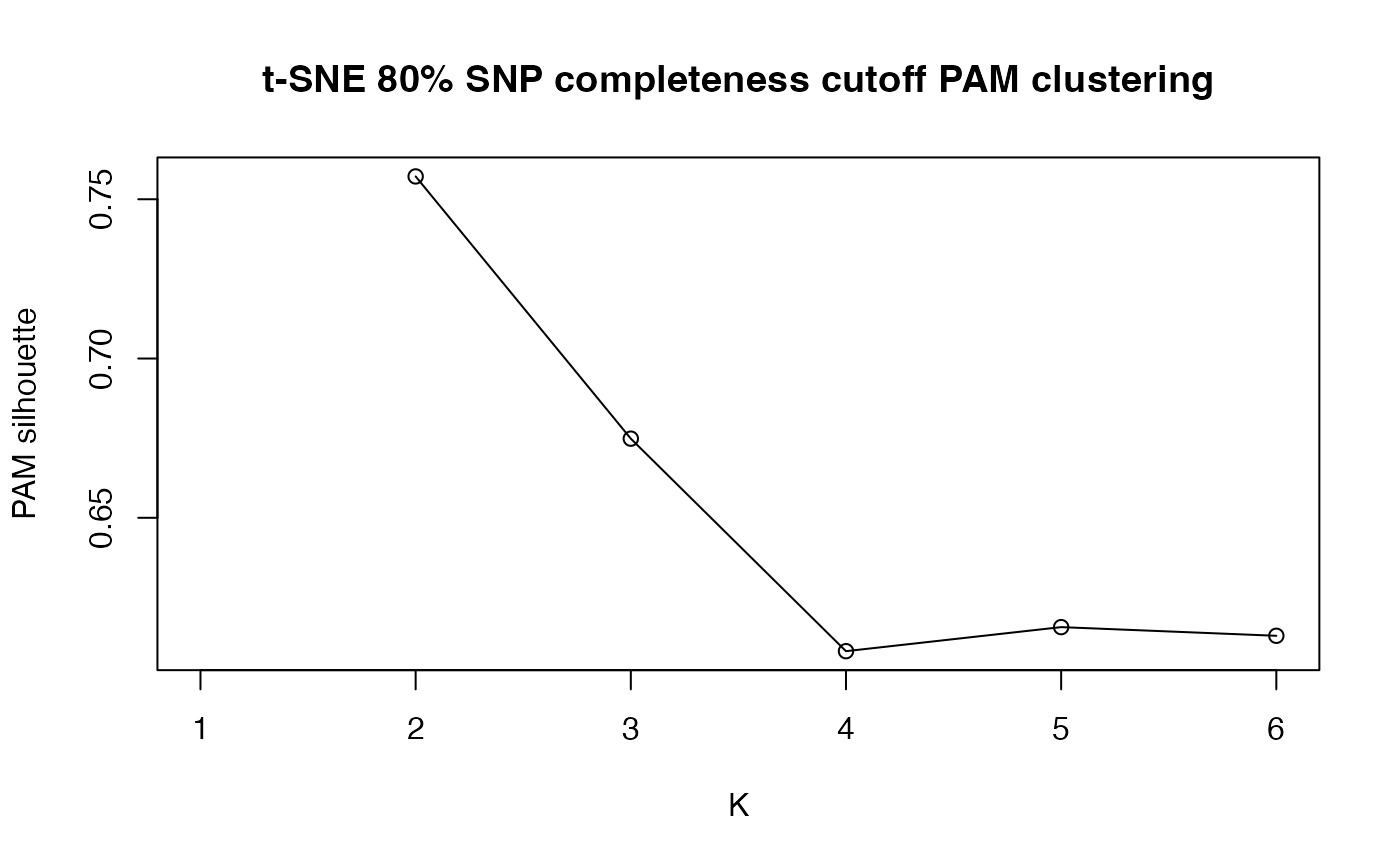
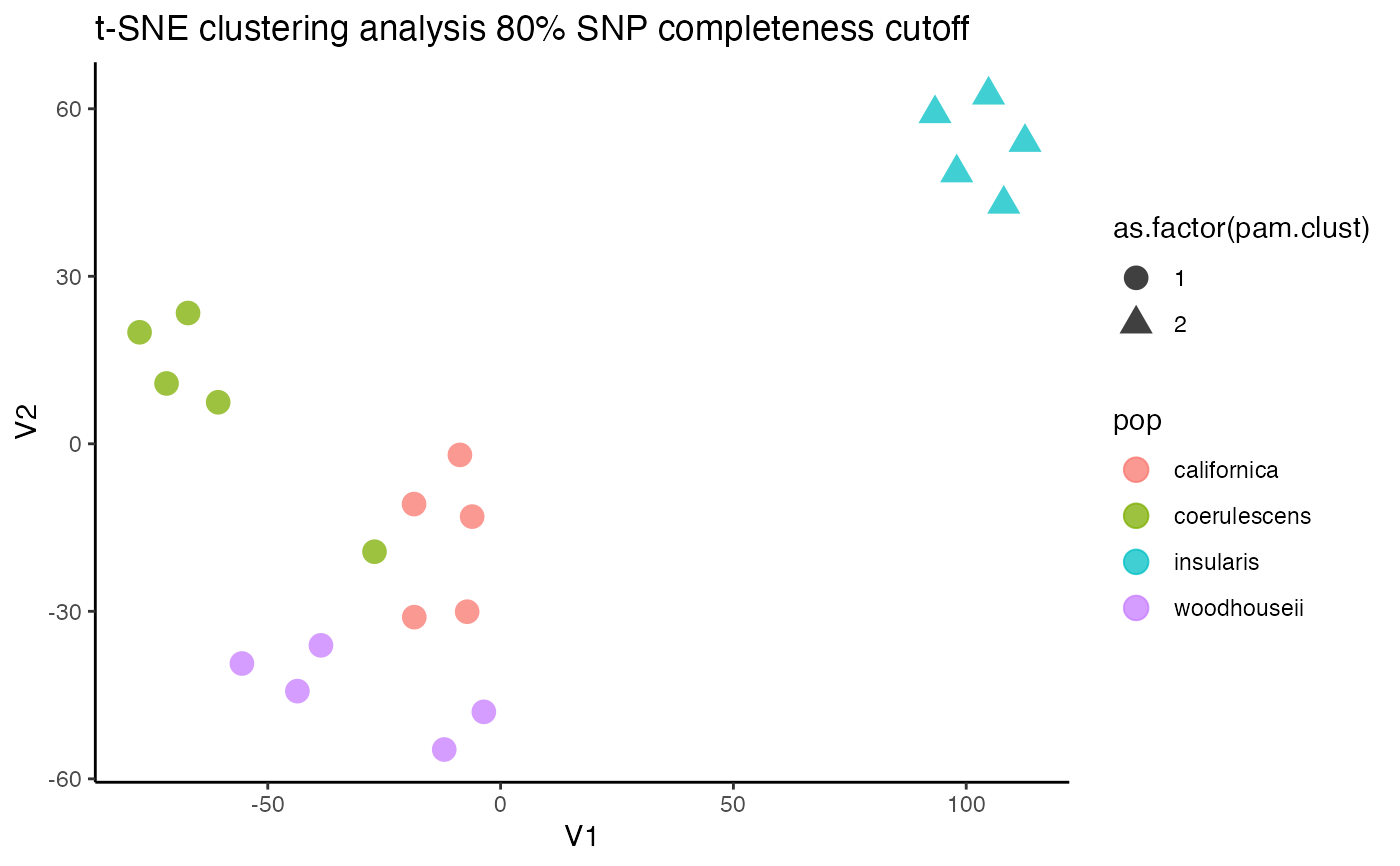
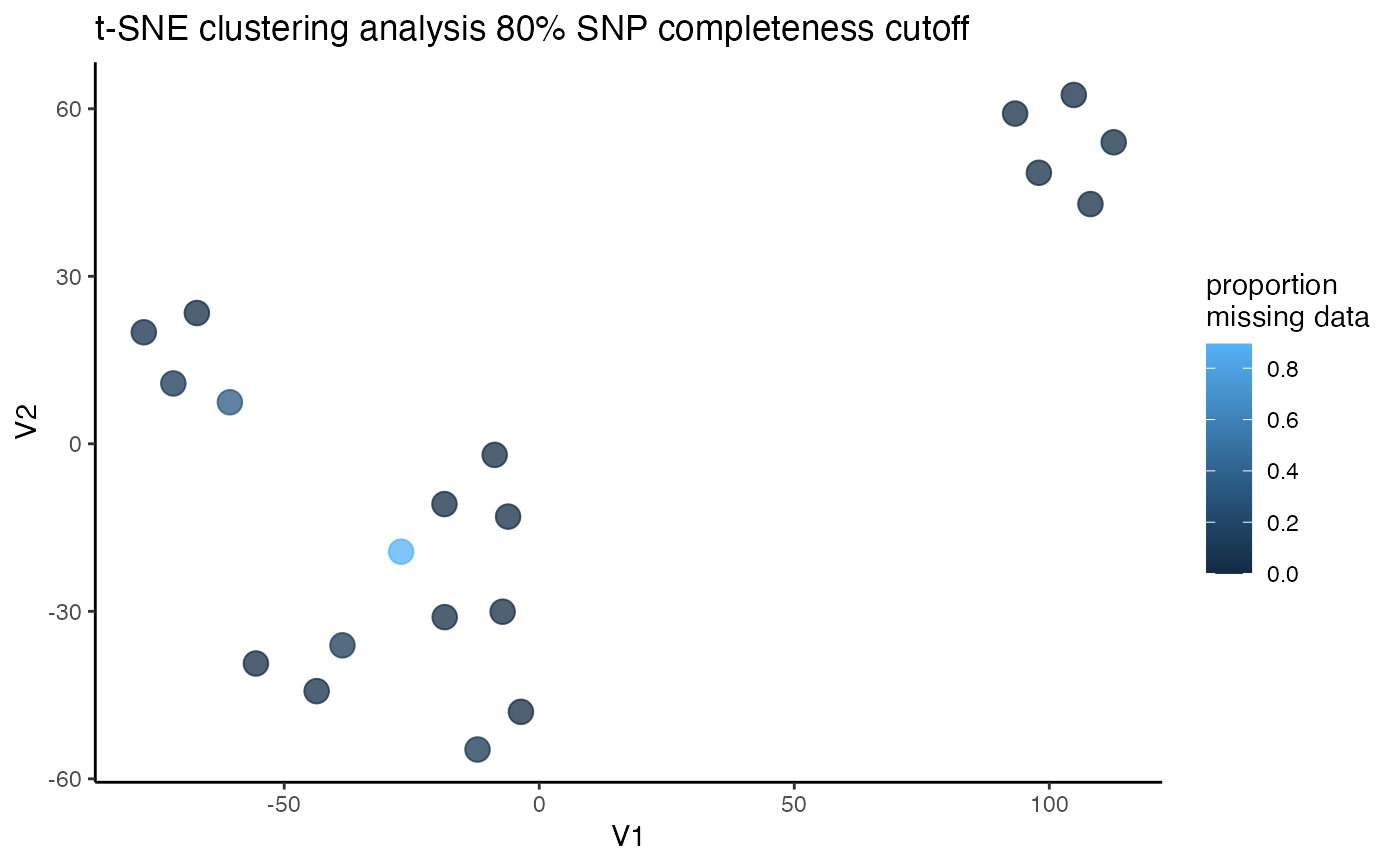 #> [[1]]
#> V1 V2 pop pam.clust missing
#> 1 -7.181015 -30.067725 californica 1 0.005847953
#> 2 -8.737704 -1.984322 californica 1 0.005847953
#> 3 -18.535683 -31.043675 californica 1 0.017543860
#> 4 -18.584513 -10.795087 californica 1 0.000000000
#> 5 -6.106049 -13.036955 californica 1 0.005847953
#> 6 97.957443 48.538775 insularis 2 0.005847953
#> 7 104.804006 62.472450 insularis 2 0.000000000
#> 8 108.065762 42.935323 insularis 2 0.005847953
#> 9 112.624444 54.000094 insularis 2 0.005847953
#> 10 93.297023 59.141590 insularis 2 0.000000000
#> 11 -3.595212 -48.000726 woodhouseii 1 0.000000000
#> 12 -12.102731 -54.750675 woodhouseii 1 0.076023392
#> 13 -55.552616 -39.344629 woodhouseii 1 0.000000000
#> 14 -38.583519 -36.090752 woodhouseii 1 0.116959064
#> 15 -43.640929 -44.292812 woodhouseii 1 0.011695906
#> 16 -77.530603 19.973745 coerulescens 1 0.029239766
#> 17 -60.653381 7.440146 coerulescens 1 0.327485380
#> 18 -27.077862 -19.325383 coerulescens 1 0.894736842
#> 19 -71.741261 10.808866 coerulescens 1 0.087719298
#> 20 -67.125596 23.421752 coerulescens 1 0.005847953
#>
#> [[1]]
#> V1 V2 pop pam.clust missing
#> 1 -7.181015 -30.067725 californica 1 0.005847953
#> 2 -8.737704 -1.984322 californica 1 0.005847953
#> 3 -18.535683 -31.043675 californica 1 0.017543860
#> 4 -18.584513 -10.795087 californica 1 0.000000000
#> 5 -6.106049 -13.036955 californica 1 0.005847953
#> 6 97.957443 48.538775 insularis 2 0.005847953
#> 7 104.804006 62.472450 insularis 2 0.000000000
#> 8 108.065762 42.935323 insularis 2 0.005847953
#> 9 112.624444 54.000094 insularis 2 0.005847953
#> 10 93.297023 59.141590 insularis 2 0.000000000
#> 11 -3.595212 -48.000726 woodhouseii 1 0.000000000
#> 12 -12.102731 -54.750675 woodhouseii 1 0.076023392
#> 13 -55.552616 -39.344629 woodhouseii 1 0.000000000
#> 14 -38.583519 -36.090752 woodhouseii 1 0.116959064
#> 15 -43.640929 -44.292812 woodhouseii 1 0.011695906
#> 16 -77.530603 19.973745 coerulescens 1 0.029239766
#> 17 -60.653381 7.440146 coerulescens 1 0.327485380
#> 18 -27.077862 -19.325383 coerulescens 1 0.894736842
#> 19 -71.741261 10.808866 coerulescens 1 0.087719298
#> 20 -67.125596 23.421752 coerulescens 1 0.005847953
#>